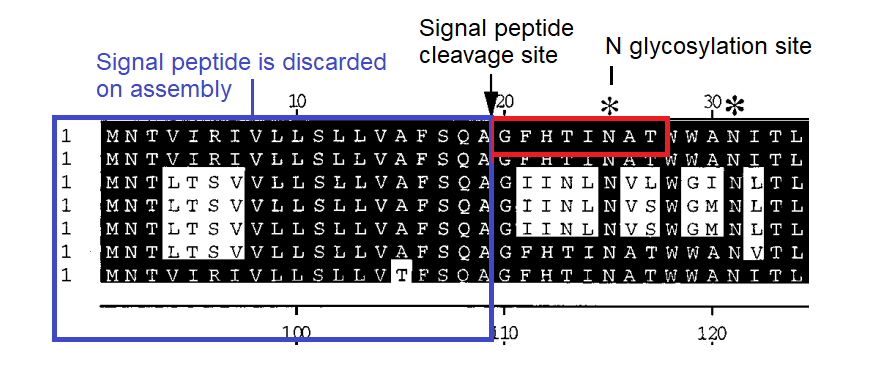
Uncanny Inserts in the N-Terminal Domain
Early in the pandemic IIT scientists Pradhan et al attracted controversy when they identified sites in SARS-CoV-2 as inserts from HIV. I propose alternative explanations that are equally "uncanny"
***NOTE to subscribers - I’m moving my articles to a new home at covorigin.org as Elon Musk’s feud with Substack has caused my traffic from Twitter to drop substantially. I’m new to Substack and my subscriber base is small (but highly valued!) so I’m reliant on that Twitter traffic. I’ll keep cross-posting for a while to see how it gets resolved, but there may be occasional glitches.
The Pradhan et al controversy marked the end of any semblance of reasonable discussion about pandemic origins and seems to have been the trigger for the infamous Fauci/Farrar teleconference. After this a lab origin became a “conspiracy theory” and a concerted effort was made to suppress discussion of the idea. Pradhan et al admitted their pre-print was written hastily, and perhaps didn’t get everything right. But they, quite sensibly, wanted to alert the world urgently to sequence anomalies they thought important for understanding pathogenesis and the possibility of an artificial origin.
They were criticized mostly because the regions of identity they found are small, and the corresponding regions in HIV are variable. HIV’s variable loops mutate even during the course of an infection within one individual (which is potentially decades long). So there are many possible HIV sequences these could match, and there wasn’t a single genome or region that was the source of their matches, but rather different regions in different genomes. Their hypothesis was perhaps over-reach, but it always came with caveats and disclosure.
But there are some things about which they were correct, that were lost in the shameful censorship and mockery that followed:
These are all inserts compared to sequences known at the time. The only viruses having any similarity to those regions were the PLA published sequences ZC45 and ZXC21. However, since that time new sequences (starting with RaTG13) have been published that tend to obscure this fact, and, I suspect, intentionally.
The regions do have homology with HIV-1’s variable loop regions, not only in the linear sequence, but structural and post-translational (e.g. N-glycosylation sites). This could also have suggested a natural explanation – convergent evolution. Sadly even this discussion was suppressed.
Even if sequence identity to HIV-1 was coincidental it might have pointed to important viral functions (e.g. co-receptor binding, trafficking by immune system cells via DC-SIGN/L-SIGN, mechanisms of immune evasion via evolving patterns of glycosylation) and aided the development of safe vaccines and therapeutics.
Regardless of their origin, these sites have always been of interest to researchers. They have indeed proved to be evolutionary hotspots in the formation of new immune evasive variants, and there is evidence to suggest a possible role in co-receptor and/or sialic acid binding. Somewhat serendipitously I’ve found some surprising, but perhaps more plausible, explanations for their origin – in conserved regions of various distantly related human coronaviruses.
Assessing the probability of a sequence occuring “at random”
Prior to SARS-CoV-2 there were 6 known human coronaviruses (229E, OC43, NL63, HKU1, MERS, SARS). Each has a spike of ~1250 aas for a total of 8750 amino acids. This is small dataset, so finding a small sequence of common amino acids is within it is more significant than finding it within a variable HIV region (many thousands of sequences), let alone finding it in an unrestricted blast search of Genbank (which contains >10 trillion bases). It isn’t unexpected to find a simple 5 amino acid match between SARS-CoV-2 and another hCoV at random (we’d expect to find about one every 366 positions), in each case there are circumstances that make it far more unlikely, but calculating probabilities somewhat more difficult. Further complicating any discussion on probability is that evolution isn’t random, and also that all of these inserts occur at positions that are functionally important. So any numbers mentioned are just to give a rough idea.
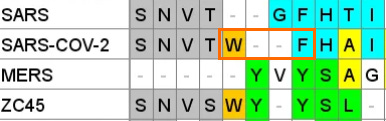
NTD Loop 1: A mash-up of Adenovirus D and MERS?
I featured this region in the article on SARS-1, because it seems almost identical to the first few residues of the NTD of certain types (e.g. 37, 19) of an Adenovirus D protein found to be associated with sialic acid binding causing broad tissue tropism resulting in diverse clinical symptoms including conjunctivitis, corneal infection and pharyngitis.
Interestingly the significance of this protein was featured in a paper published just a few months before the first SARS case in 2002.
In SARS-1 the homology to Adenovirus 37 is striking, and improbable. It’s simple to calculate there are 25.6 billion possible combinations of 8 amino acids. A 7 out of 8 match is more frequent but still around 1 in 200 million. And that’s without the additional stipulation of a conserved N-glycosylation site in the final residues.
In SARS-CoV-2 some residues from the Adenovirus sequence remain, but there is also interesting homology to MERS, a region identified as forming part of its sialic acid binding pocket. The homology was first identified in early 2020 by a University of Warwick team):
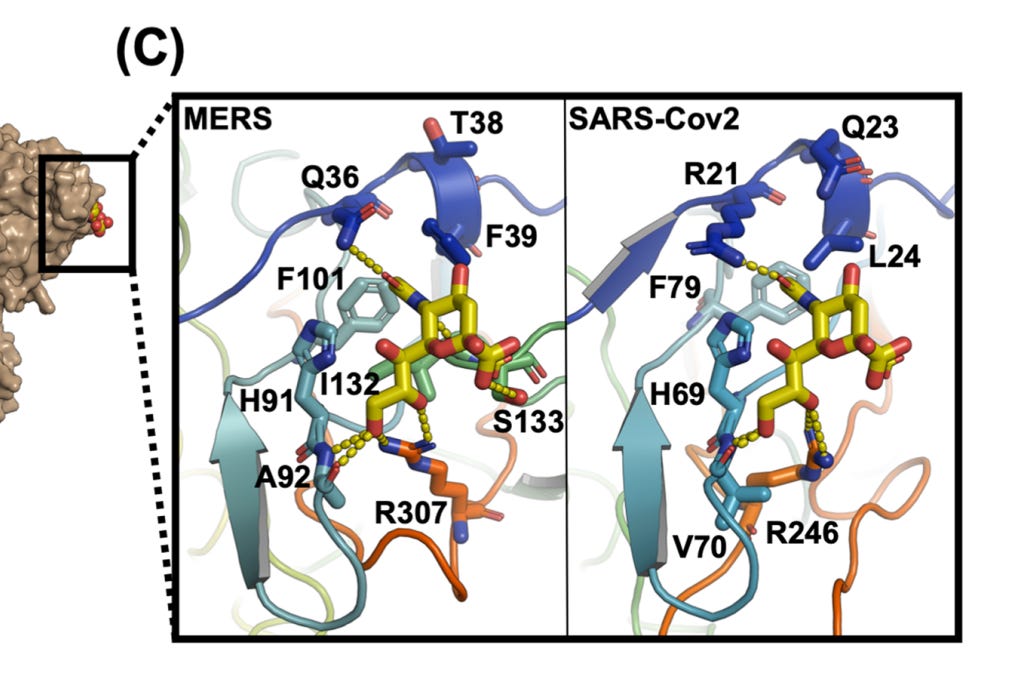
A paper from June 2019 paper (Tortorici et al). found a conserved sequence in coronaviruses that have been previously confirmed to bind sialic acids: OC43, BCoV, PHEV and HKU1. The sequence is different in MERS, but two aromatic amino acids – Tryptophan
adjacent to a Phenylalanine (F) – occupy similar structural positions, despite being separate in the linear sequence. At the base of this loop in SARS-CoV-2 an adjacent W-F pair is formed, that is not present in MERS.
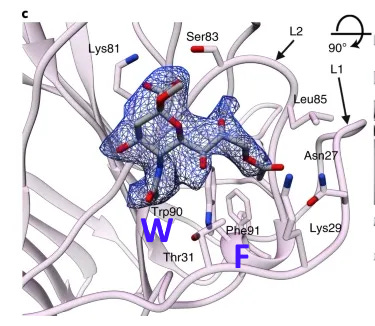
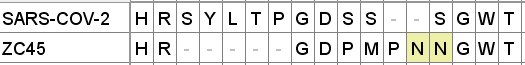
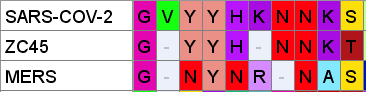
A curious pattern in ZC45
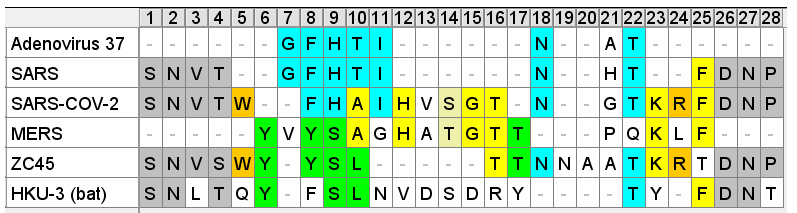
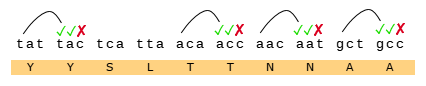
This Tryptophan residue is one of a couple shared uniquely with ZC45/ZXC21 at this site. These sequences are of interest for their provenance (being published by a largely PLA team from Nanjing Command in 2018). Notice also in ZC45 at this site the unusual number of doublets YY-SL-TT–NN–AA and the pattern of nucleotides, with the wobble base varying in each member of a pair.
Although the homology between ZC45 and SARS-CoV-2 isn’t exceptionally strong at this particular site, its significance is best understood in context of the other sites in the NTD, and some uniquely share mutations elsewhere in the genome (such as E and S2). Viewed in totality these suggest ZC45 maybe also an engineered virus (or fabricated sequence). This has important implications regarding the intentions of the engineers, suggesting a premeditated release of the virus, with ZC45/ZXC21 intended to provide evidence that such mutations had a precedent in nature.
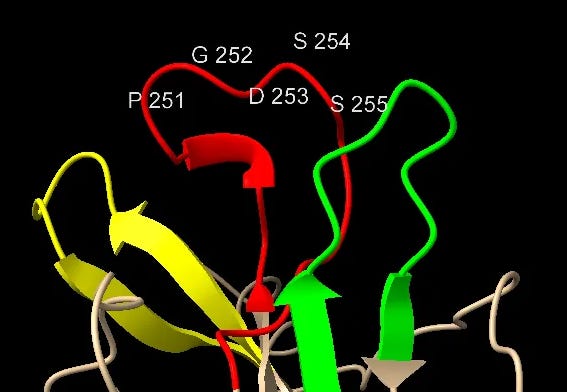
NTD Loop 3: An ACE-2 binding region from NL63
*Numbering reflects position within the gene sequence, not the article.
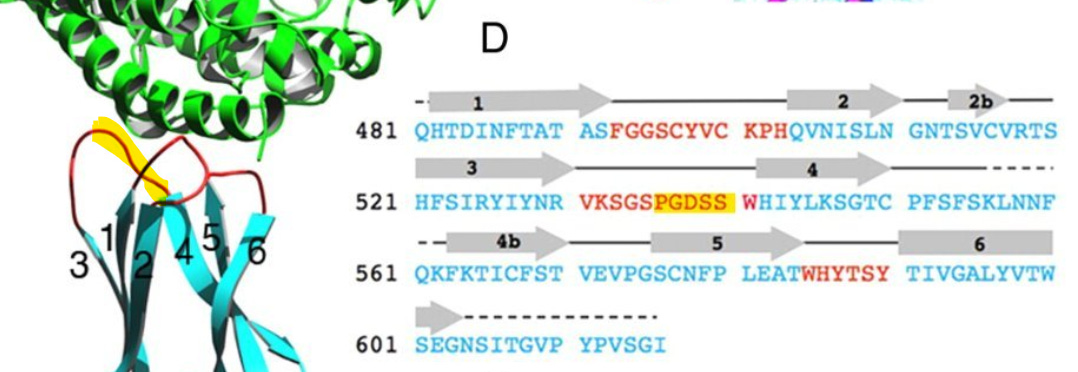
This sequence is another extended exterior loop and also is an insert compared to related bat viruses. This sequence contains a 5 amino acid sequence PGDSS which is identical to one from unrelated human coronavirus NL63 – which as with SARS and SARS-CoV-2 uses ACE2 as its main receptor.
Once again there is also homology with ZC45/ZXC21, particularly in flanking residues, that is not seen in SARS or other bat SARS-like viruses known at the time.
But it isn’t just the 5 aas of sequence identity, 13 out of 14 residues in this region match, but in a different position, (including one conservative substitution (R→K). In a loop region which is intrinsically disordered, and contains many flexible residues (such as Serine (S) and Glycine (G)) the order in which they occur is likely less important.

Most important is the location of this sequence in NL63. According to a 2009 paper (Wu et. al.), which includes Fang Li as senior author, NL63’s RBM is composed of three loops separate in the sequence but adjacent in 3D structure. The SARS-CoV-2 NTD site matches the central loop of these (highlighted below). It’s also interesting that in SARS-CoV-2, three NTD loops that are separate in the sequence, come together in a similar way in the 3D structure.
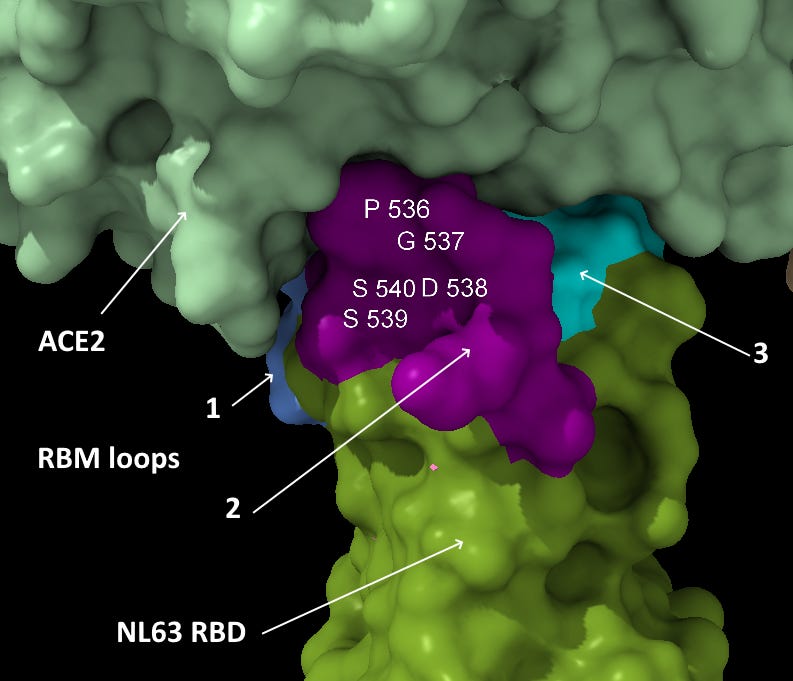
This NTD loop also has homology with MERS (identified by Baker et al) in the residues at each end of the loop.
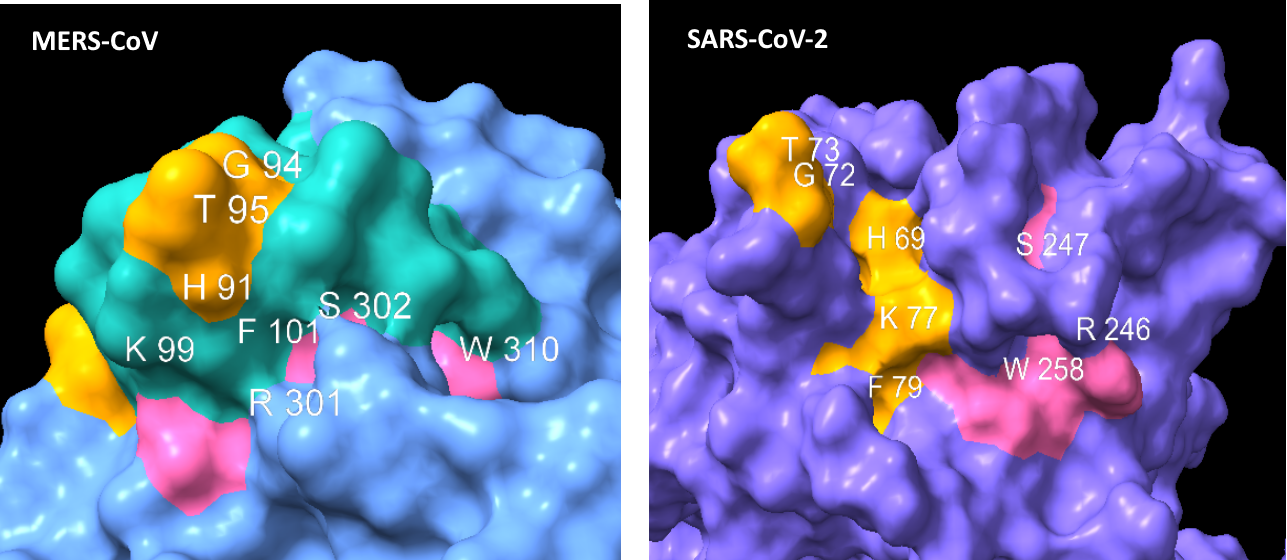
Though it is only 3 residues at this site it’s important to look at the 3D structural context, Sialic acid binds in a concave surface or “pocket”, the position of the active residues relative to the surface topology is crucial.
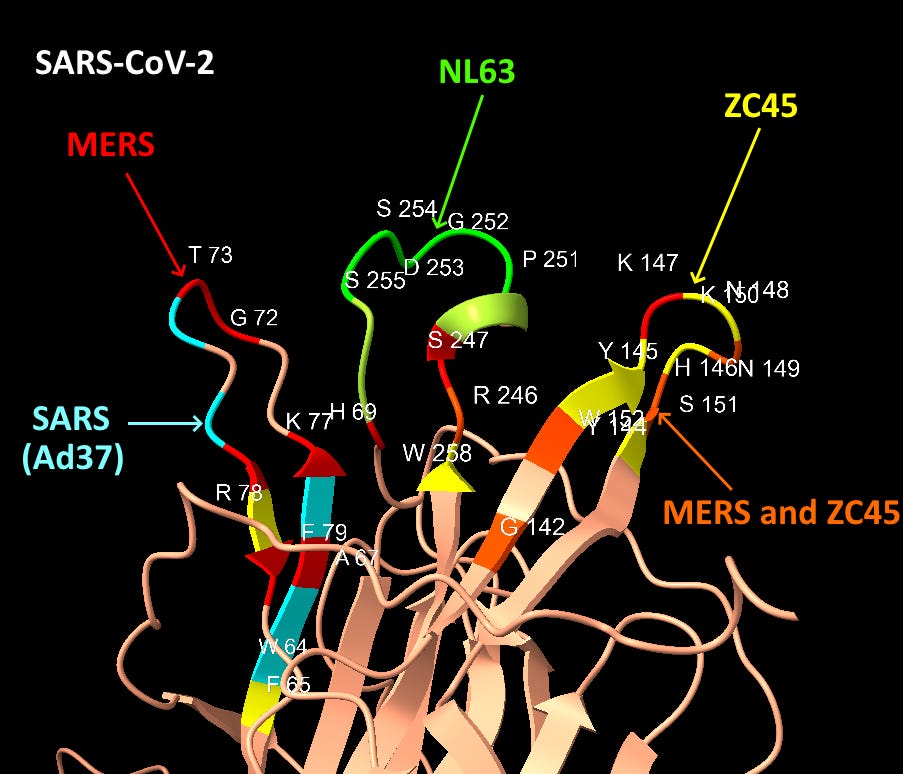
Looking first at Loop 1, the common residues between MERS and SAS-CoV-2 in this loop are coloured gold. The similarities here are obvious, in each case we have a G and T protruding like a finger, and an H, K and F in a recessed region at the base.
Though less easy to make out in a still image, the S, R and W residues from Loop 3 (coloured pink) also occupy structurally similar positions in the binding pocket (although in SARS-CoV-2 case the surface formed is concave but flat, not a hollow groove as in MERS).
NTD Loop 2: ZC45
If our middle loop is derived from NL63, and the green loop is a SARS/MERS mash-up (via Adenovirus D), could the third loop also have homology to other human CoVs?
In this case the sequence aligns rather well with ZC45 – purportedly a bat, not human virus. The substitution S→T towards the 3’ is conservative and the N-glycosylation site is conserved
But, as I previously said, I have doubt that ZC45 is itself a natural virus, so could ZC45 have obtained this insert from elsewhere? The 4-mer sequence NNKS occurs in SARS-1 NTD at a different position. Just downstream is another NN pair which also creates a potential N-glycosylation site (or choice of):
Another curious coincidence I noticed in ZC45 is that all of the three corresponding NTD loops in its sequence contain an Asparagine pair NN and two contain a Tyrosine pair YY
I speculate that these sequences in ZC45 are experimental constructs based on observations made from the NTD of SARS-1. In this instance SARS-CoV-2’s corresponding region looks like a small refinement, whereas the others have been more radically reworked. But again there is some homology to MERS sequence in a structurally proximal loop, including the N-glycosylation site
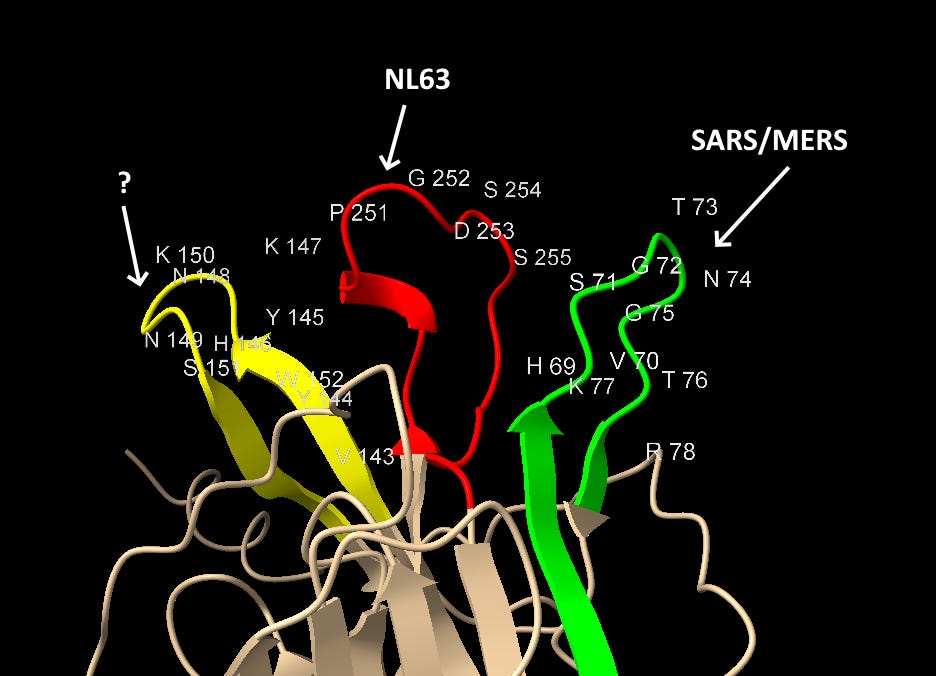
Graphical Summary of NTD Loop Homology
The structures of SARS-CoV-2 and MERS are compared below with identical residues highlighted in red. SARS-CoV-2 contains many important residues from MERS’ sialic acid binding structure, in three different regions of the linear sequence, but coming together in the 3D structure.
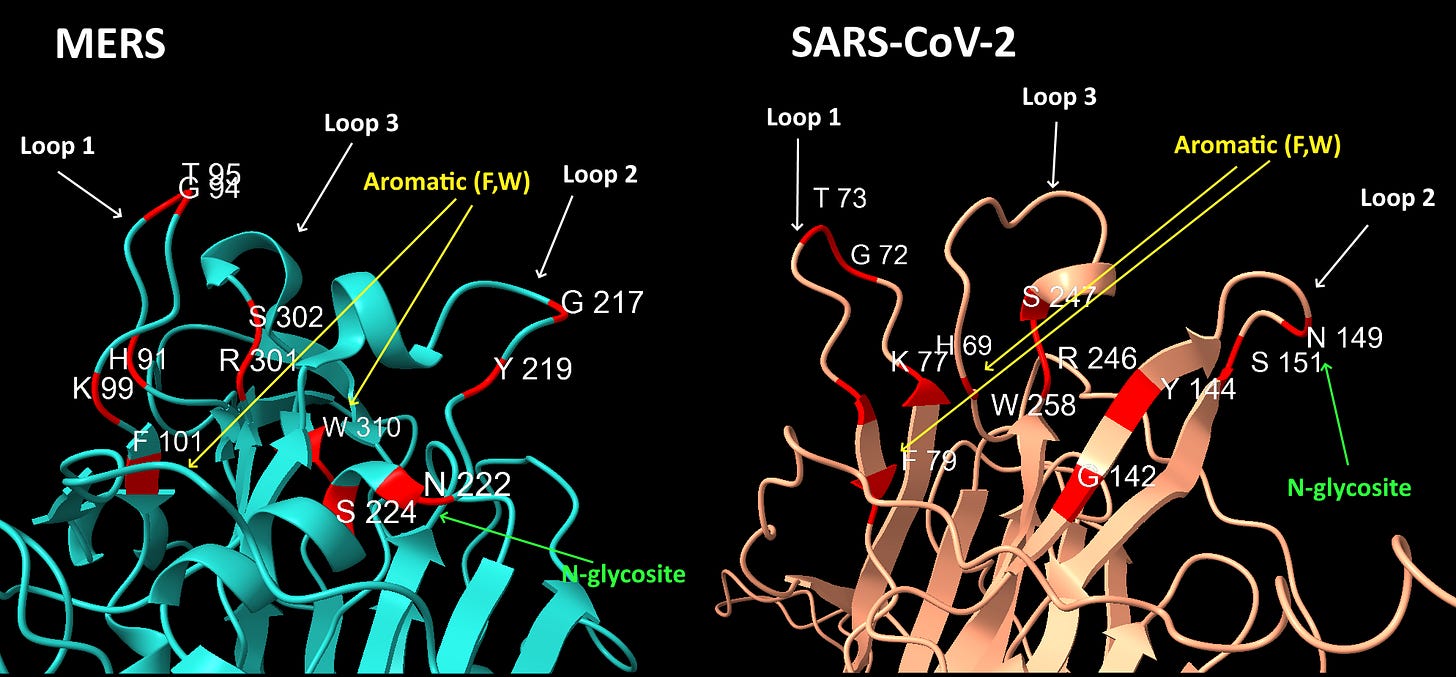
Interspersed with the MERS residues, SARS-CoV-2 also has homology with other human coronaviruses SARS and NL63, and claimed bat coronaviruses ZC45/ZXC21. The NL63 homology is particularly unexpected, as the virus is unrelated, and the sequence comes from a different domain of the spike.
Discussion
While Pradhan et al may have found the wrong source for these inserts, they correctly identified suspicious locations in the SARS-CoV-2 spike. There are now many sequences that have been published since the start of the pandemic obfuscating the origin of these inserts, but also many reasons to ignore these as possible fabrications. I’ll substantiate this further in future articles.
I occasionally receive criticism that the kind of engineering needed sounds overly complex, but I’d point out that a grant application from Lanying Du states the following in the abstract:
“We use the RBDs from highly pathogenic coronaviruses, including MERS-CoV and SARS coronavirus (SARS-CoV), as the model system...we will construct chimeric RBDs containing the core subdomain from one coronavirus RBD as the structural scaffold and the RBM from another coronavirus RBD as the immunogenic sites”
Almost exactly as SARS-CoV-2’s NTD appears to be. More about this soon.







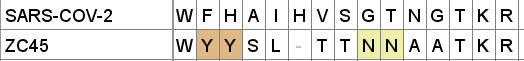
I recently found new evidence that ZC45 and ZXC21 may have a dubious origin. The NCBI's sequence read archive has an "Analysis" tab which shows a breakdown of what percentage of reads are estimated to come from different organisms. It was implemented using a software called STAT which searches for matches to 32-base k-mers. The database which contains the results of the STAT k-mer analysis is about 500 GB in size, but it can be searched through BigQuery at the Google Cloud Platform: https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud-based-taxonomy-analysis-table/.
I used BigQuery to search for runs which had over a thousand k-mer matches to sarbecoviruses: ```select * from `nih-sra-datastore.sra.metadata` as m, `nih-sra-datastore.sra_tax_analysis_tool.tax_analysis` as tax where m.acc=tax.acc and tax_id=2697049 and total_count>1000 order by releasedate```. I found 384 runs from 74 unique studies that were older than the first SARS 2 runs from January 2020: https://cdn.discordapp.com/attachments/1093243194231246934/1108414826352484383/bigquery-sarbecovirus-1k.json.
The most interesting result was a metagenomic sequencing run which matched ZXC21 and which seems to have been overlooked earlier by COVID researchers: https://www.ncbi.nlm.nih.gov/sra/?term=SRR5351760. It was titled "metagenomic sequencing of bat metagenome:zhoushan intestine", it was submitted by "Research Institute for Medicine of Nanjing Command", and the samples are supposed to have been collected between July and August 2015, which matches the collection date of ZC45 and ZXC21 at GenBank. And the study of the run was by the same authors who published the paper about ZC45 and ZXC21.
When I aligned the reads in the SRA run against a FASTA file of sarbecoviruses, I got 78% coverage for ZXC21 with an average error rate of 0.8%, I got 16% coverage for ZC45 with an average error rate of 1.6%, and I got below 3% coverage for all other viruses (so I'm not sure if the sample contained both ZXC21 and ZC45 or only ZXC21, since some reads from ZXC21 probably also matched ZC45):
curl https://ftp.ncbi.nlm.nih.gov/entrez/entrezdirect/install-edirect.sh|sh
esearch -db nuccore -query '(sarbecovirus "complete genome" NOT txid2697049) OR NC_045512.2'|efetch -format fasta>sarbe.fa # `NOT txid2697049` excludes SARS 2 and `OR NC_045512.2` includes Wuhan-Hu-1
brew install seqkit bowtie2 samtools gnu-sed
wget ftp://ftp.sra.ebi.ac.uk/vol1/fastq/SRR535/000/SRR5351760/SRR5351760_{1,2}.fastq.gz # download from ENA because it's faster than downloading from NCBI
seqkit fx2tab sarbe.fa|grep -Ev 'RfGB02|Rs7907'|sed $'s/A*\t$//'|seqkit tab2fx>sarbe2.fa # remove poly(A) tails, omit Rs7907 which has a segment of mammalian rRNA at the beginning, and omit RfGB02 which has an assembly error at the end
bowtie2-build --threads 3 sarbe2.fa{,}
bowtie2 -p3 -x sarbe2.fa -1 SRR5351760_1.fastq.gz -2 SRR5351760_2.fastq.gz --no-unal|samtools sort -@2 ->SRR5351760.bam
samtools coverage SRR5351760.bam|awk \$4|cut -f1,3-6|(gsed -u '1s/$/\terr%\tname/;q';sort -rnk4|awk -F\\t -v OFS=\\t 'NR==FNR{a[$1]=$2;next}{print$0,sprintf("%.2f",a[$1])}' <(samtools view SRR5351760.bam|awk -F\\t '{x=$3;n[x]++;len[x]+=length($10);sub(/.*NM:i:/,"");mis[x]+=$1}END{for(i in n)print i"\t"100*mis[i]/len[i]}') -|awk -F\\t 'NR==FNR{a[$1]=$2;next}{print$0"\t"a[$1]}' <(seqkit seq -n sarbe2.fa|gsed 's/ /\t/;s/, .*//') -)|column -ts$'\t'
There's also a little-known public API endpoint which shows the number of K-mers for an SRA run which match each species and each taxonomical node: https://trace.ncbi.nlm.nih.gov/Traces/sra-db-be/. It shows that for the run SRR5351760 which matched ZXC21, there's a total of 415,912 assigned K-mers, but only 14,488 of them were assigned to bacteria and only 74 to the order of Enterobacterales, and there's zero k-mers assigned to Escherichia coli or to the genus Escherichia:
curl 'https://trace.ncbi.nlm.nih.gov/Traces/sra-db-be/run_taxonomy?acc=SRR5351760&cluster_name=public' >SRR5351760.stat
jq -r '.[]|.tax_table|sort_by(-.total_count)[]|(.total_count|tostring)+";"+.org+";"+(.tax_id|tostring)+";"+(.parent|tostring)' SRR5351760.stat
The metagenomic sequencing run for RaTG13 is also supposed to represent a "fecal swab", but it's inconsistent with how the sample contains very few reads which match intestinal bacteria. See this preprint by Steve Massey: https://arxiv.org/abs/2111.09469.
You can compare the results to the fecal metagenomic sample of RmYN02: https://trace.ncbi.nlm.nih.gov/Traces/sra-db-be/run_taxonomy?acc=SRR12464727&cluster_name=public. There's 26,684,391 identified reads out of which 18,526,075 are classified under bacteria, so it's a total of about 69% as opposed to about 3% for the ZXC21 sample or about 0.6% for the RaTG13 sample. And there's 368,425 k-mers assigned to Enterobacterales and 5,886 to Escherichia.