How the PLA Planned a Plague
Planning the cover-up of SARS-CoV-2 commenced long before the outbreak. It is key evidence SARS-CoV-2 was developed as a bioweapon - not an accident. PLA fingerprints are everywhere.

In previous articles I’ve documented the deceptive nature of WIV’s claims to have discovered bat viruses with human pandemic potential in Yunnan. Initially this seemed to be about laying a false evidentiary trail for a natural origin of SARS-1. But with the “discovery” of RaTG13 they moved on to preparing the cover-up of a future bioweapon.
Throughout their endeavors WIV were shadowed by Tu Changchun, of the AMMS Institute of Military Veterinary Medicine (IMVM). The IMVM is based in the city for which Tu is named: Changchun, capital of Jilin province, in northern China formerly known as Manchuria. Though Tu doesn’t have a high profile, he has a central and recurring role in the SARS origin saga. Wherever there’s a gap in the evolutionary evidence, Tu has an uncanny knack of being in the right place and time to sample/sequence precisely what is needed.
Of the many teams who joined the search for SARS-related coronaviruses other than WIV, only Tu Changchun’s group claimed to have found any with a SARS-like RBM i.e. one that is capable of efficiently binding human ACE2.
Tu’s discovery LyRa11 was published to GenBank around the same time as WIV’s discovery was announced in Nature in 2013. While Tu stayed out of the limelight, Lyra11 provided necessary corroboration of WIV’s discoveries - which have flaws pointing to their artificial origin.
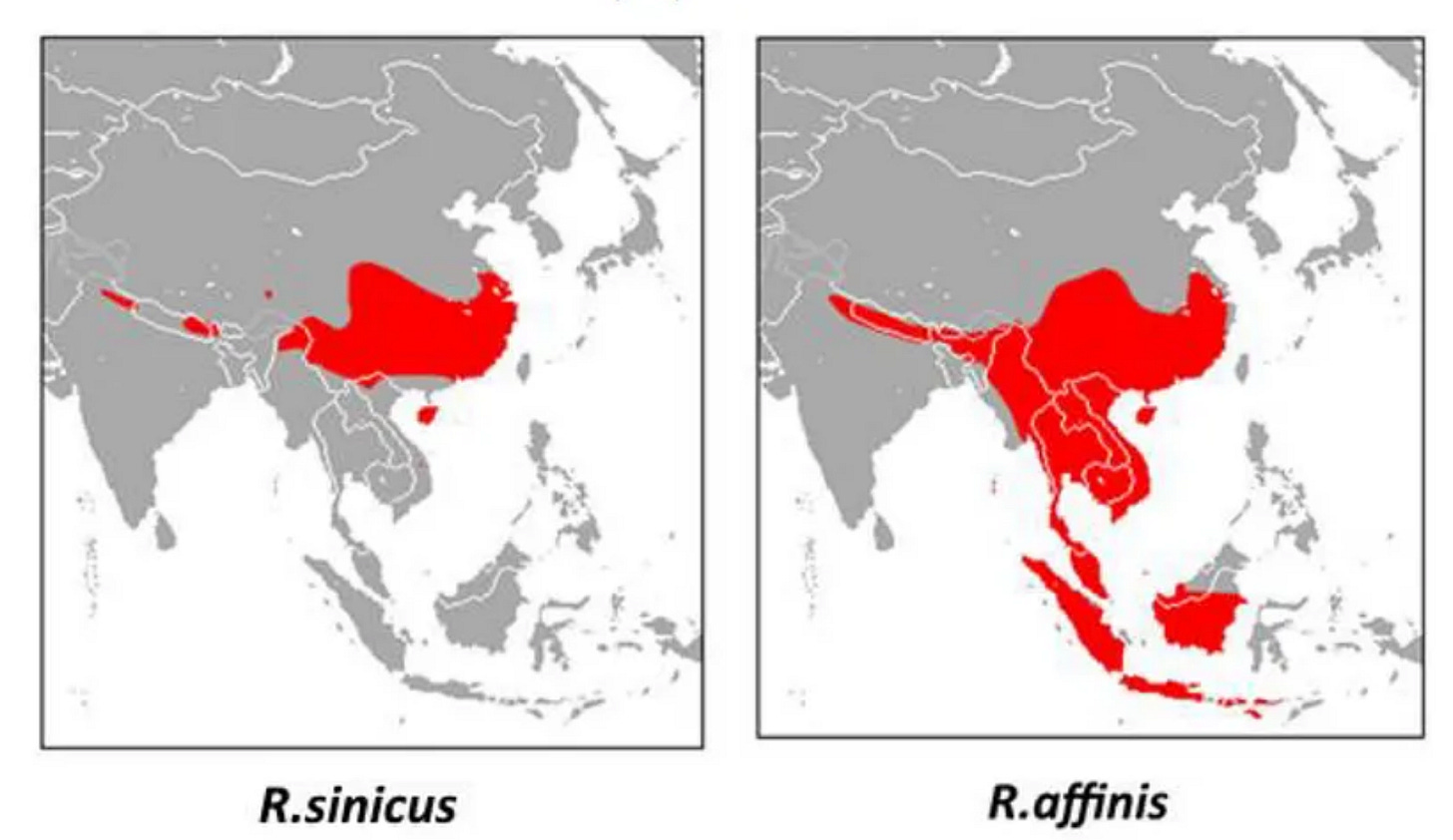
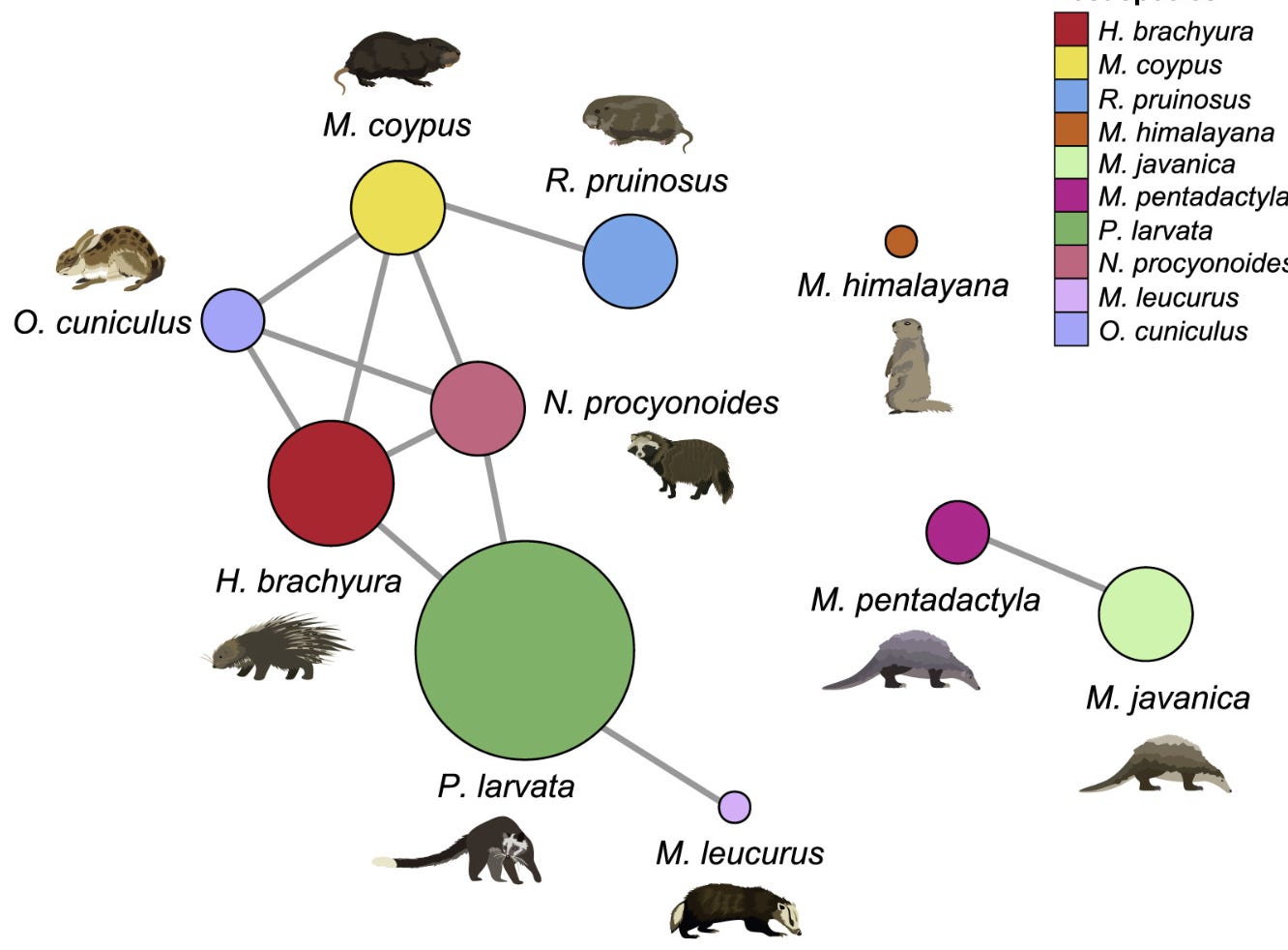
LyRa11 was purportedly found on the other side of Yunnan, near the town of Baoshan. Baoshan was the site of a horrific Japanese biological warfare attack in May 1942. LyRa11 was sampled from a different horseshoe bat species: R. affinis, whereas WIV’s “discoveries” had all been in R. sinicus. At that time R. affinis wasn’t known to host any SARS-related virus, but soon it would become known as the host of RaTG13. The significance of the host species is that R. affinis has a range that extends deep into South-East Asia, while R. sinicus prefers cooler climes in China. Due to host species, and a more southerly site, RaTG13 is within range of Manis javanica, the Malayan pangolin.
Early in the pandemic, the recombination of a Malayan pangolin coronavirus and a RaTG13-like bat virus was, for a time, the preferred explanation of natural evolution of SARS-CoV-2. But the evolutionary puzzle was still missing some pieces.
ZC45 and suppressed sequences
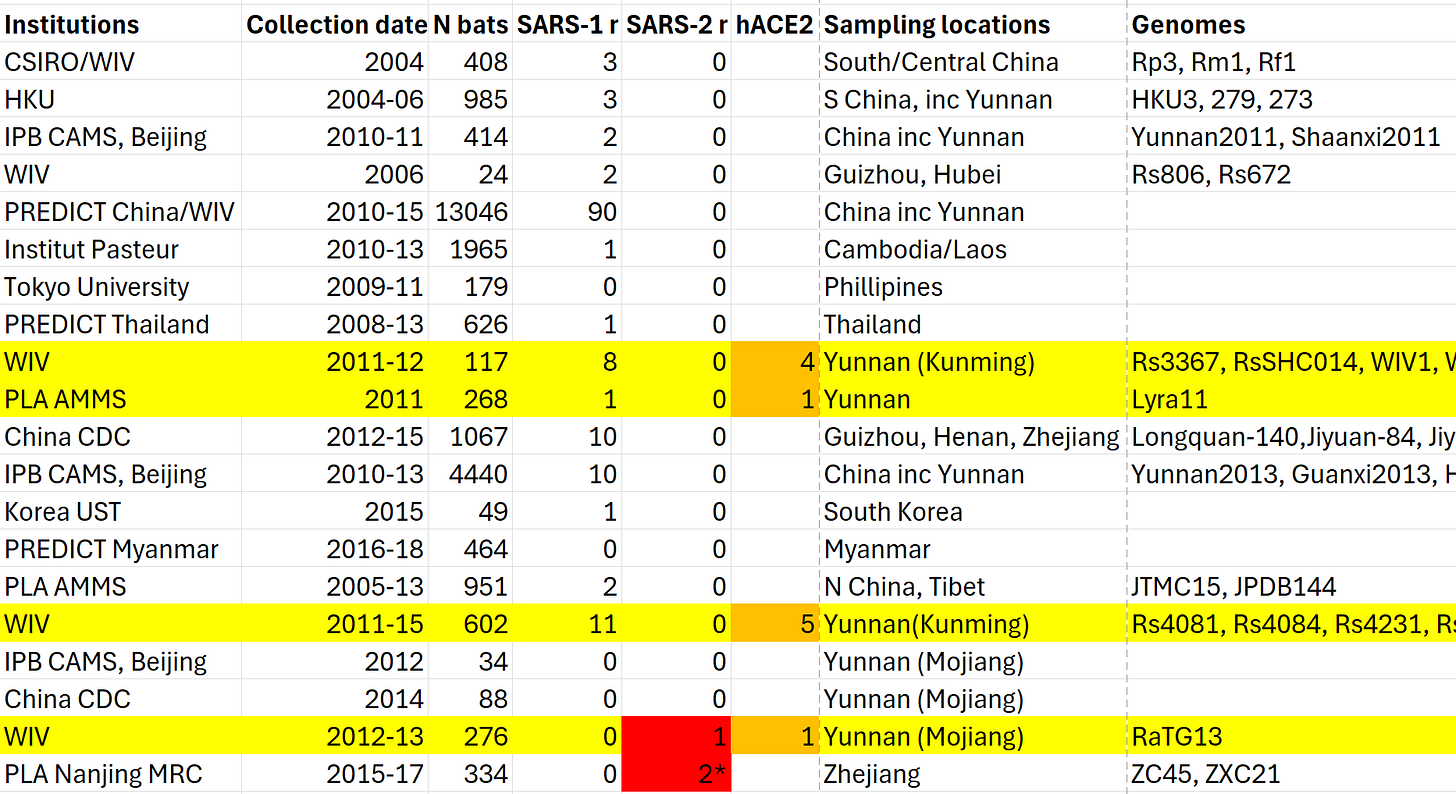
ZC45 was the first member of the SARS-CoV-2 clade to be published, by a group from the PLA’s Nanjing Command Medical Research Center. It was published on March, 28th, 2018, coincidentally (perhaps) the day after a consortium including Ralph Baric, Linfa Wang, Zhengli Shi, Peter Daszak submitted the notorious DEFUSE proposal. In the proposal, Baric stated they wanted to find viruses approximately 20% different to SARS. ZC45 is 19.4% different. What’s more, its PLA discoverers had already shown it could infect suckling mice. It would have made an ideal case study for DEFUSE.
Curiously, WIV also had viruses with spike sequences 99% similar to ZC45. These were purportedly found in Guangdong province, some 1200km away. But WIV kept these hidden from the public, and don’t seem to have disclosed them to their DEFUSE collaborators either. They uploaded them to GenBank in July 2018 but kept them in suppressed mode. They were briefly made visible when a pre-set embargo expired in July 2022, but they immediately asked NCBI to hide them again.
These ZC45-like sequences don’t seem genuine. There are far too few amino acid differences between them for viruses that evolved so far apart, but there are many silent mutations. The abnormal ratio of these types of mutations is a sign of likely fraud. Also their RdRp sequences place them nowhere near ZC45 in the phylogenetic tree. What game were WIV playing?
Unlikely recombination
ZC45 has an interesting relationship with the coronavirus purportedly sampled from sick pangolins captured in Guangdong (MP789). There is very high homology in the spike NTD (green). But in the RBD, ZC45 is more like a genuine SARS-1 related bat virus (e.g. Rp3), while MP789 is suspiciously close to SARS-CoV-2.
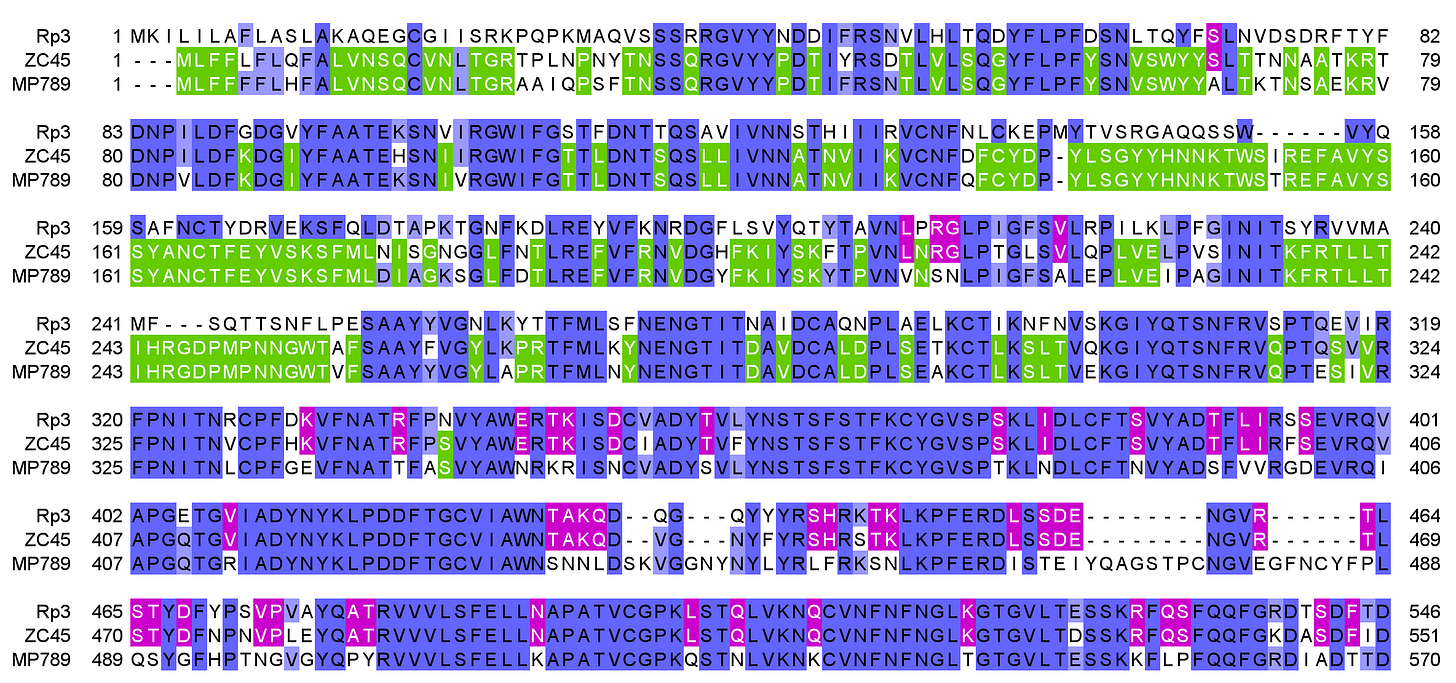
The NTD is usually one of the most variable parts of the spike and genome as a whole. It has surface loops that are tolerant of mutation, and it's highly exposed to the immune system at the tip of the spike. Given also that these two viruses were sampled far apart and in different host species, we should expect many more amino acid differences between them.
ZC45 was found in Zhoushan, an archipelago off the port of Ningbo, in Zhejiang province. It was hosted by R. Pusillus - which is known as the least horseshoe bat as it measures just 3-4cm in length and flies only short distances.
MP789 was sampled in Guangdong, from pangolins seized from smugglers and held in a wildlife rescue center. Some scientists suggested the pangolins may have been infected at their origin in South-East Asia, or by the smugglers, or other animals they were smuggled alongside. This may have deviated from the original script prepared by the PLA.
On January 20th, Shi Zhengli submitted a paper proposing an origin based around a ZC45-like virus - while omitting to mention that she had a much closer sequence (RaTG13) or that she had ZC45-like sequences sampled in Guangdong. This was no mistake, but more deception. To what end?
Just two days later pangolin data was uploaded to GenBank, and WIV submitted the pre-print about RaTG13, a virus far closer to SARS-CoV-2 (if it is genuine). Attention was drawn away from ZC45.
“In war, no plan survives first contact with the enemy”.
On 19th January, with indications an epidemic was spreading uncontrolled in Wuhan, HKU virologist Dr Li-Meng Yan appeared on a popular Chinese language livestream to publicize the PLA connection to ZC45, and the official cover-up of transmission between humans. This brought an immediate response. China admitted to human transmissibility the next day. Three days later a harsh lockdown was imposed on Wuhan.
With these events, some started talking about the outbreak becoming “China’s Chernobyl”, a prediction it might lead to the communist regime’s demise, as had happened to the USSR. Connections to the PLA, had to be obfuscated.
WIV’s lab was commandeered by the AMMS. Some believe Shi Zhengli erratic disclosures were made under duress, or that she had defied orders and been punished. Having investigated her past work, I doubt she was so heroic. But with whistleblowers being punished and threatened, it should have been obvious that it wasn’t an environment conducive to open independent science. Data about the origin should never have been trusted.
PLA fingerprints on pangolin data
In 2022, Adrian Jones, Daoyu Zhang et al took a deep dive into pangolin sequencing data. Some of this data had been shared between multiple groups, and its provenance remains unclear. Jones et al looked at all available datasets, used in several different papers, including a more recent one with pangolins sampled after the outbreak.
They found:
Very little RNA from pangolin coronavirus overall, and only in a handful of samples. There were more reads from non-pangolin viruses.
Only 9 coronavirus reads in total covering the RBM (the key link to SARS-CoV-2).
Insufficient read coverage to assemble the complete MP789 genome.
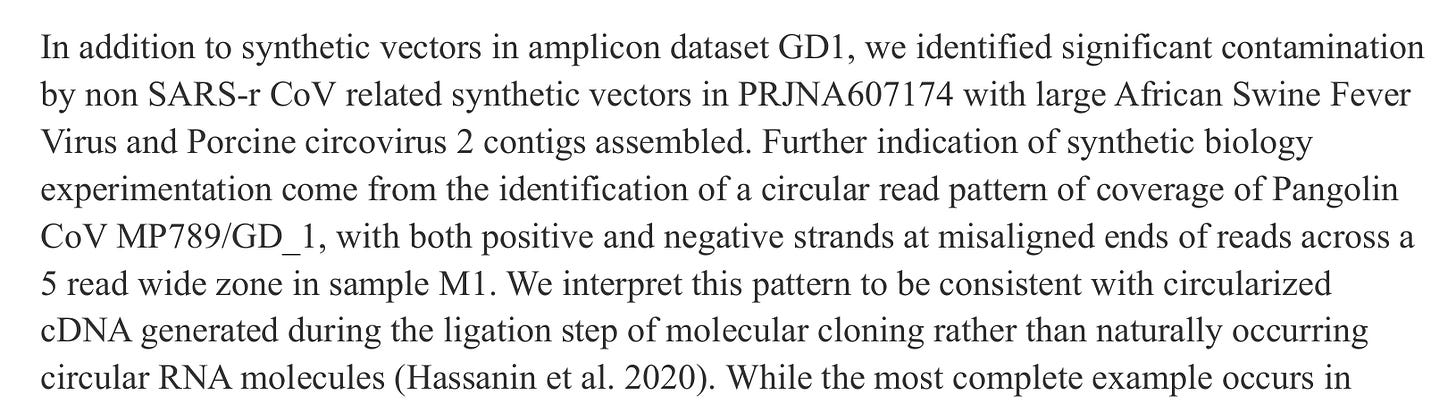
Contigs with highly structured repeats at each end, suggestive of cloning in a circular plasmid.

Reads that matched synthetic DNA cloning vectors.
In the more recent dataset, reads from an unpublished coronavirus closely related to ZC45, which was not identified by the paper’s authors.
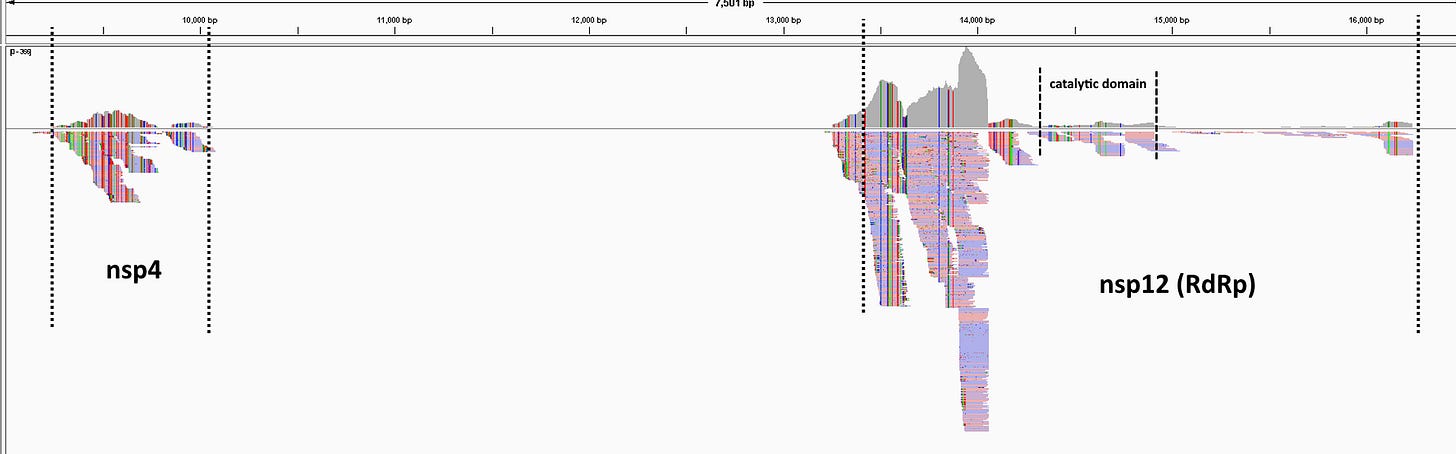
Novel virus - or evidence of artifice?
While Jones et al did some excellent analysis, I disagree with their conclusion these reads came from an unpublished coronavirus.
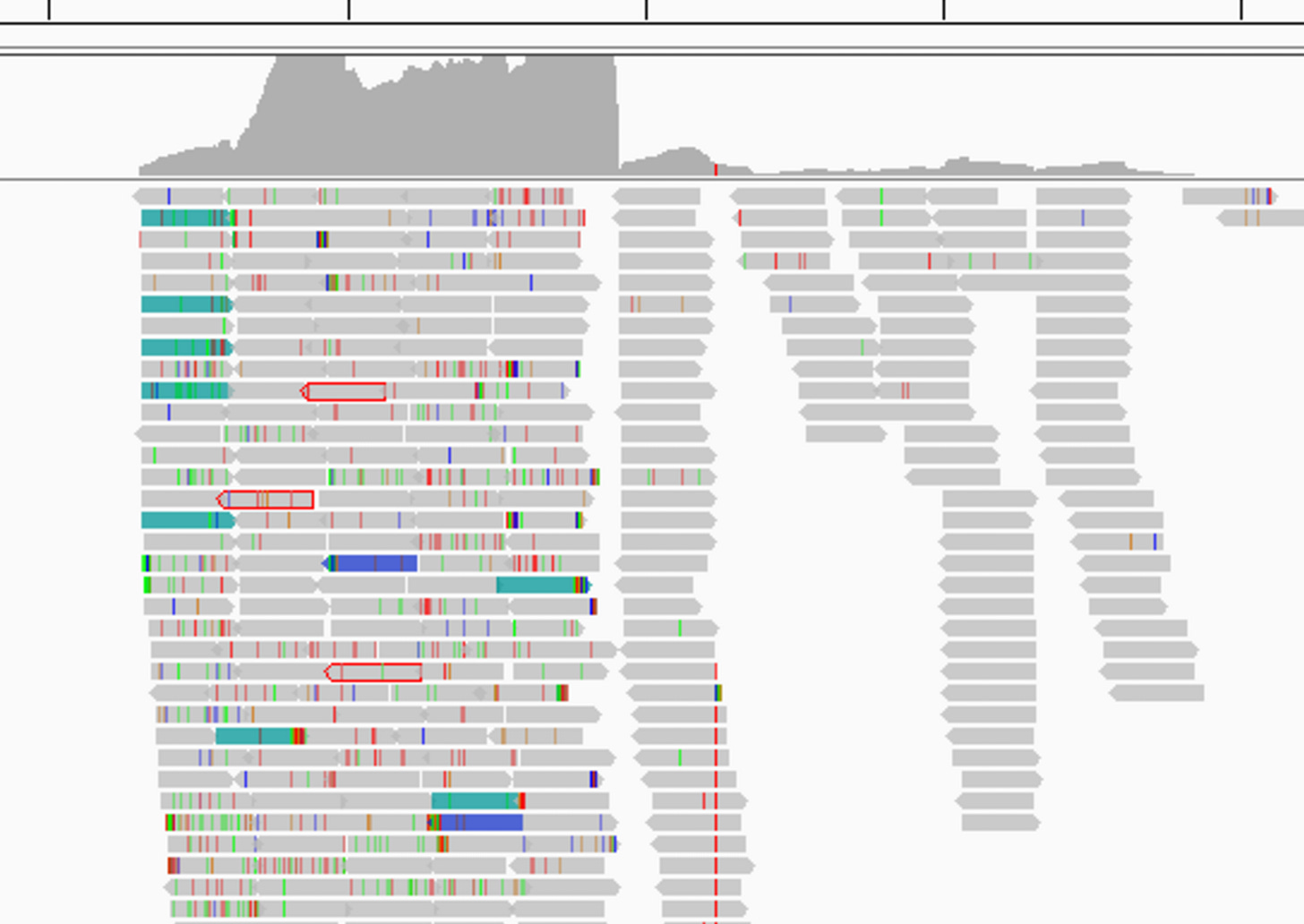
The reads together cover only ~9% of a coronavirus genome, and read depth is very uneven, high over one end of the RdRp, but much lower over the catalytic core (typically the most conserved region, commonly used as a “genetic fingerprint”). There is a large gap with no coverage between nsp4 and the RdRp. Jones et al point out that while an assembled sequence aligns best to ZC45 in the RdRp, it is closer to the Guanxi (GX) pangolin coronavirus in the nsp4 region. N.B. GX is a different pangolin virus to the GD virus discussed above. There is low homology between them in some regions (e.g. S1).
There is far too much genetic diversity between the individual reads for them to have come from a single virus. Some reads in the RdRp have 100% identity to ZC45, but many are ~95% identical, and some are as low as 75%. Blasting those with lower identity shows them to be closer to other coronaviruses e.g. HKU3, Anlong-89, SARS-1, than they are to ZC45.
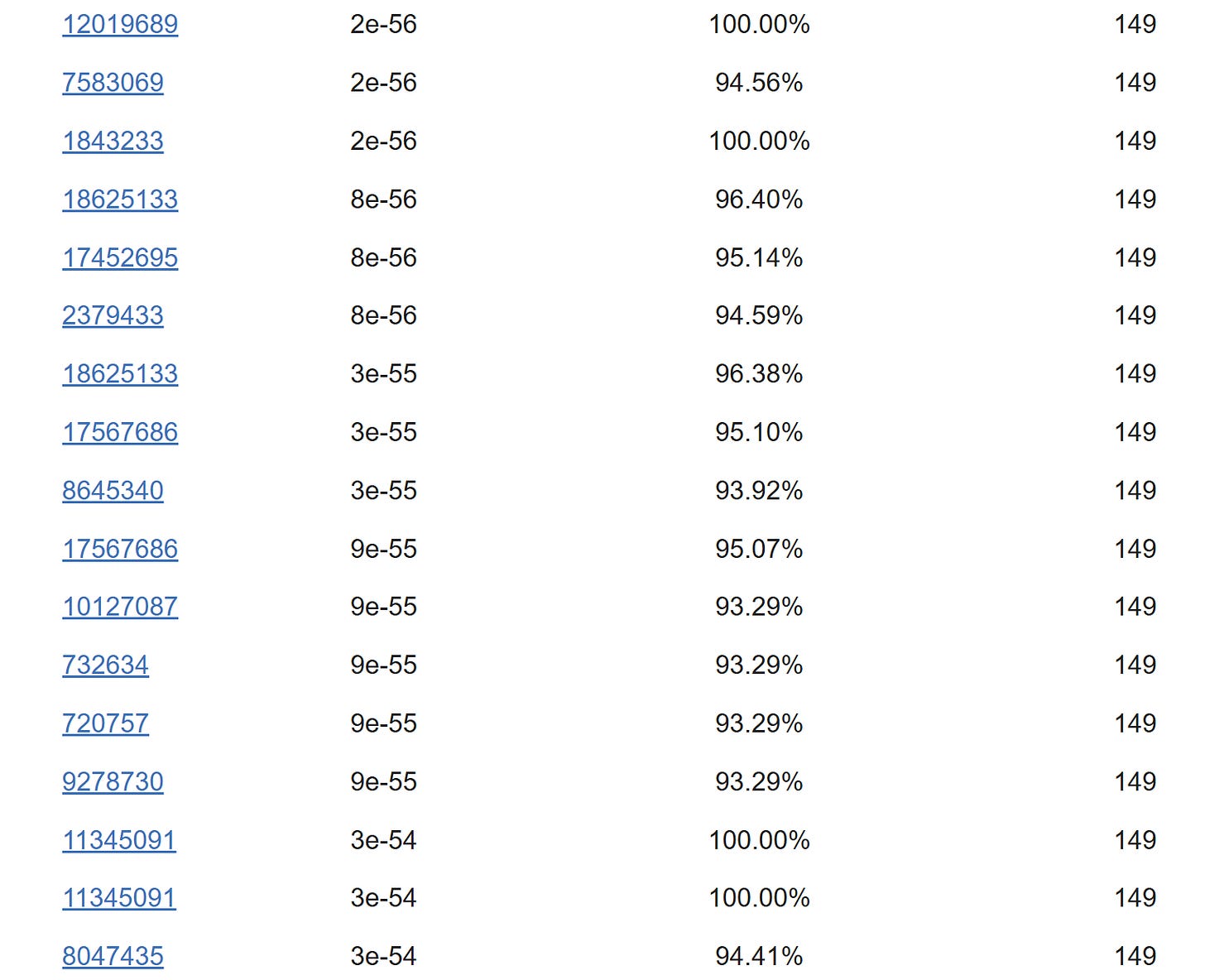
Of the reads in nsp4 the highest is ~98% identical to the GX pangolin coronavirus, but only ~85% identical to ZC45. Again, there is too much genetic diversity for them to have come from a single virus.
My interpretation is that this is evidence of genetic engineering - a directed evolution experiment intended to generate random chimeric sequences. This was likely done with techniques such as DNA shuffling and error prone PCR as described in a previous article. The nsp4 reads are likely from a similar but separate experiment. The purpose of these experiments could be to support fraudulent studies on coronavirus evolution, distribution, host switching - in which often only the RdRp is used. Alternatively, they may have been intended to become part of novel viral genomes, assembled from individually optimized genes.
Whose fingerprints are on the data?
Jones et al highlighted several other contaminating viruses that might offer clues to the provenance of the datasets. Among others, they identified African Swine Fever Virus and Porcine circovirus type 2.
Porcine viruses like African Swine Fever are commonly studied by agricultural universities in China, but also by the IMVM, particularly in 2018-19.
I found the Porcine circovirus 2 reads noted by Jones et al, to be from an unpublished species which is less than 80% identical to any species in GenBank. Finding novel circovirus genotypes was a focus for IMVM during 2017-19, with Tu Changchun’s involvement.
In a subsequent paper, Jones et al found a novel MERS-related virus in reads from a different dataset published by a similar group of authors. The authors claimed the dataset was derived from pangolin samples, but Jones et al determined it was from wild boar. Tu Changchun and IMVM also surveyed wild boar in 2017-19.
Some of the sequencing data was uploaded by Nanjing Agricultural University (NJAU). According to documents obtained by Colonel Lawrence Sellin, NJAU virologist Shuo Su has been funded by the IMVM. The director of IMVM, General Xia Xianzhu, is an alumnus of NJAU. It seems likely that the pangolin samples and/or read data were provided to the group via Tu Changchun. Tong Yi-Gang of AMMS Beijing Institute of Microbiology and Epidemiology (BIME) claims to have isolated and sequenced the GX pangolin virus in 2017, his group may be the original source.
The pangolin trail leads back to Zhejiang
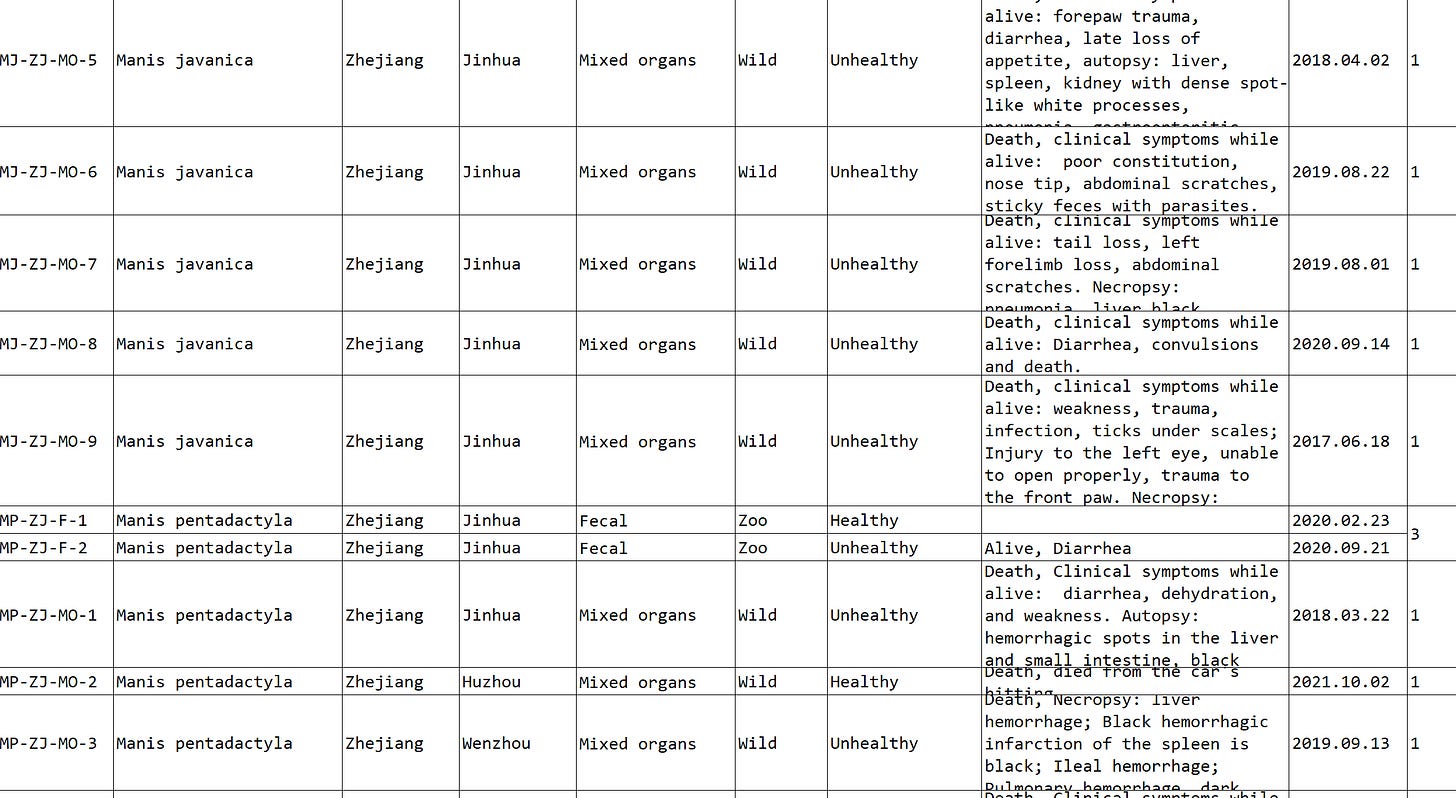
Interestingly, the Manis Javanica pangolin samples analyzed in this paper were all said to have been found in Jinhua, Zhejiang province (~200km from Zhoushan) in the wild. Although there is a Chinese pangolin species (Manis pentadactyla) which is native to the region, it is far from the range of Malayan pangolins. How did they get there?
In June 2019, an article appeared in an international conservation magazine about the practice of releasing seized smuggled Malayan pangolins into the wild. Some local authorities were apparently unable to distinguish between the species.
This in turn cites a Chinese article from October 2018, which discusses specific cases of this occurring. One such incident apparently occurred in Zhejiang, but similar incidents were also reported in Yunnan and Guangdong.
The article also mentions the smuggled pangolins from which Tong Yi-gang’s group claimed to have isolated the GX pangolin coronavirus.
The “Game Animals” paper used data from both Malayan and Chinese pangolin (M. pentadactyla) samples - all collected in Zhejiang.
The paper infers that the two species can pass pathogens between each other. This seems unlikely to happen in the wild as they are both solitary species. Chinese pangolins are rare, and released Malayan pangolin rarer still.
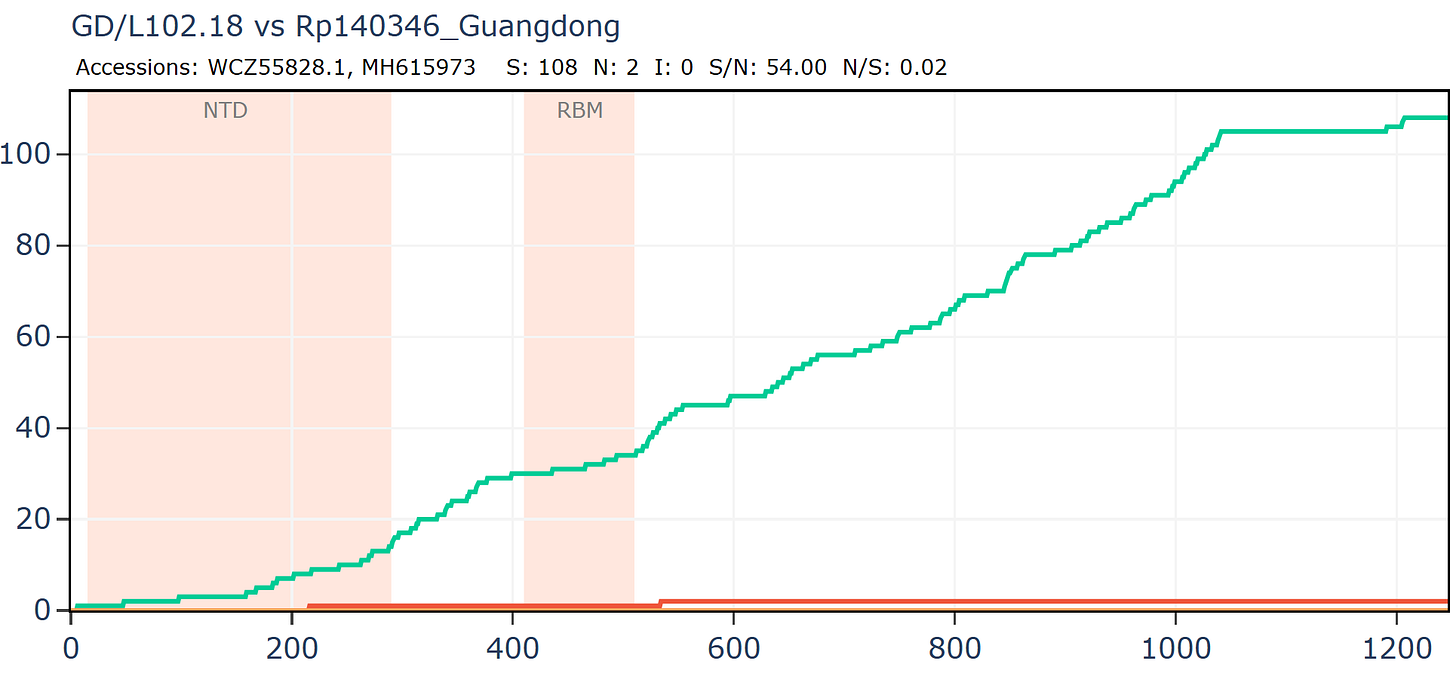
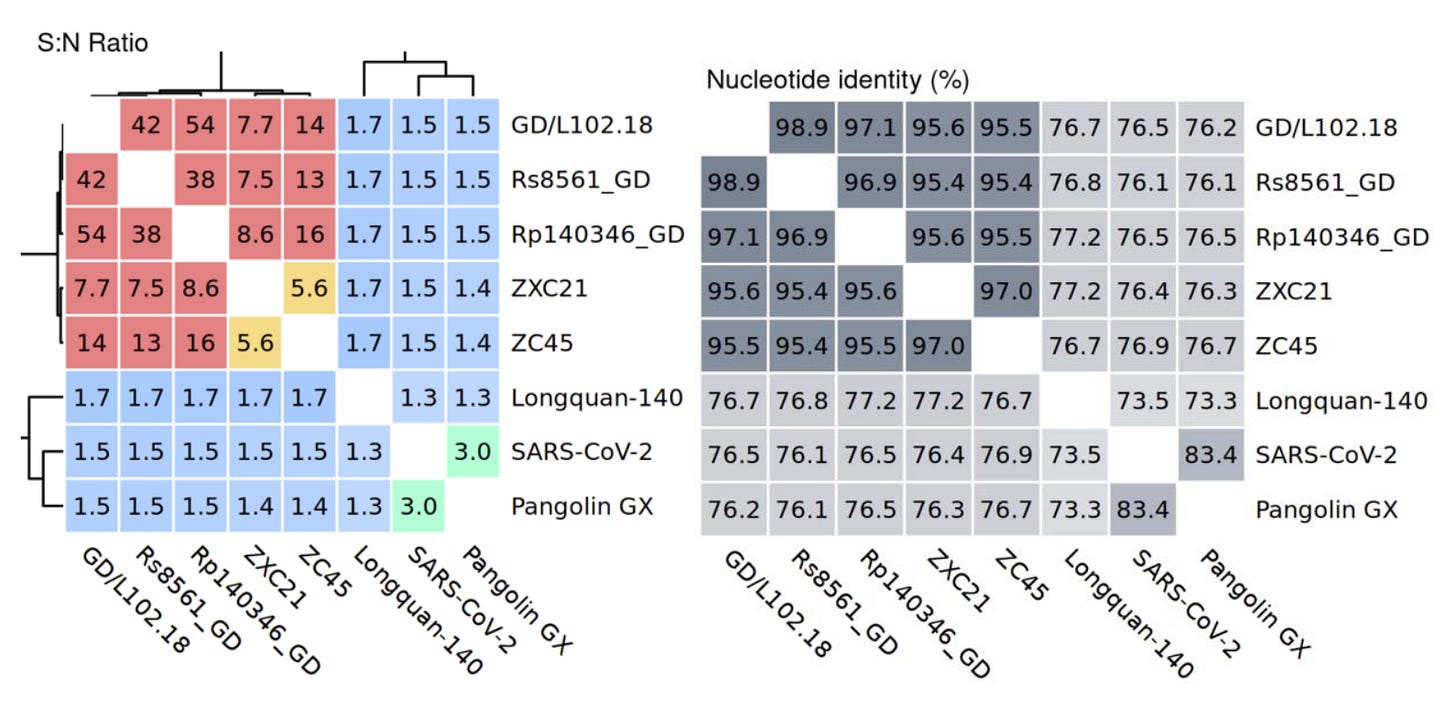
The group also published a new ZC45-like bat coronavirus from Guangdong (GD/L102.18). It has 108 synonymous mutations, but only 2 amino acid differences in the spike compared to one of WIV’s secret Guangdong sequences. This is the most extreme ratio of synonymous to non-synonymous mutations I’ve coming across, having compared thousands of pairs of viruses. I can confidently say these sequences aren’t genuine. But WIV’s sequences are needed to compare it to. These were still hidden at the time, it might have been assumed they would stay hidden.
PLA pangolin wrangler Tu Changchun
Before and during World War 2 the broader region formerly known as Manchuria, was occupied by Japanese colonial forces known as the Kwantung Army. It was the locale for the bioweapon experimentation and other atrocities committed by the infamous Unit 731 based in Harbin. Unit 731 wasn’t the only such unit. The city of Changchun housed Kwantung Army Unit 100, also known as the Warhorse Disease Prevention Unit. This unit produced offensive bioweapons targeting horses and livestock, some of which (e.g. glanders) could also infect humans. Experiments on human subjects (usually Chinese prisoners) were common. Post-war Changchun under the CCP retained an institution with similar focus (although China would claim for defensive purposes only) the Military Horse Health Research Institute, which later became the IMVM of the AMMS.
Tu Changchun is a protege of General Xia Xianzhu, who joined the Military Horse Health Research Institute after graduating from Nanjing Agricultural University, later becoming director of IMVM. Xia emphasizes building “Military-Civilian Fusion” links with civilian institutions, such as Harbin Veterinary Research Institute. Many of these also have a focus on zoonotic pathogens with biowarfare potential. This is partly a legacy of the Japanese colonial period. For years after World War 2 there were sporadic epidemics of diseases like plague linked to wartime Japanese bioweapon releases which persisted in environmental and animal reservoirs.
During the Korean War, the CCP claimed the US was deploying biological weapons near its border. This is unproven and likely just propaganda, but US failure to prosecute Japanese biowarfare criminals, and USAMRIID’s appropriation of Unit 731’s research data fueled suspicion and resentment. At the time the US and other western powers still had offensive biowarfare programs.

Tu Changchun’s contributions to the origin saga include:
In May 2003, he was involved in sampling civets to ascertain a connection to SARS. The paper was a collaboration with CSIRO’s Gary Crameri.
In 2004 he conducted animal experiments infecting civets with different strains of human SARS.
In 2009 he worked closely with Trevor Drew (then of the UK’s Animal and Plant Health Agency) to develop a reference lab for rabies in Changchun. In September 2018, Drew became head of CSIRO’s AAHL lab.
In August 2013, as WIV and CSIRO were publishing the first viruses with SARS-like RBM, Tu Changchun’s IMVM group published Lyra11, as evidence to support the existence of a SARS-like RBM in bats.
In 2016, Tu’s group claimed to have found a SARS-like virus with a distinctive 579-nucleotide deletion seen only twice before. Once in a late-stage human SARS patient, and once by WIV.
In April 2020, journalist Sharri Markson wrote a piece exposing the relationship between Trevor Drew and Tu Changchun. The piece lacked any incriminating substance. Drew in media interviews didn’t disclose CSIRO’s work with WIV and AMMS on SARS origin.
Later in 2020, Tu collaborated on a paper with Linfa Wang, CSIRO’s Gary Crameri, which claims to have sequenced a SARS-CoV-2 related coronavirus from Thai bats. They also claim a pangolin tested positive for SARS-CoV-2. They refer to “historical data” which shows a pangolin tested positive for SARS in May, 2003 but provide no reference for this.
Shame and striving
In an interesting coincidence the release of the smuggled Malayan pangolin in Zhejiang province happened near the city of Jinhua. This region was the site of Unit 731 bioweapon attacks, Chongshan Village, a few km from where the pangolin was released now hosts a memorial and center of research into Japanese biowarfare atrocities.

The port of Ningbo, off which the islands of Zhoushan sit, was the site of another notorious attack in which a Japanese plane dropped plague infected fleas, killing over 1500 people from disease and forcing the destruction of houses. Today the site is marked with a monument bearing a plaque which reads: “Never forget our national humiliation, strive to strengthen the nation”.
The PLA Eastern Theater Command (formerly Nanjing Command) today occupies the former site of Japanese Unit 1644 in Nanjing. Unit 1644 was responsible for deploying the bioweapons developed by Unit 731 in Zhejiang.
Symbolism is particularly important to the PLA and the CCP. The CCP’s domestic propaganda still emphasizes historical “humiliation” dealt to China, by Japan and western powers. But the age of imperialism is over. In the present zeitgeist, it’s the CCP’s bioterrorist aggression that will bring the shame.